
The multinational research project lasted about ten years, from 1990 to 2000, and for the first time produced reference data on the complete genome of the most important model plant, the thale cress (Arabidopsis thaliana). Today, an individual genome can be deciphered in just a few hours, and molecular biology has developed so rapidly. In many cases, the bottleneck of genome research is no longer the chemical analysis of the genetic material, but the subsequent data processing and evaluation. In order to eliminate this bottleneck, bioinformaticians at Forschungszentrum Jülich have developed a database that - at least for plant research - bundles and processes existing knowledge.
Existing knowledge is highly fragmented
"Many plant genomes have now been published," said Björn Usadel, project manager of the primary plant biotechnology database and bioinformatician at Forschungszentrum Jülich. "But these genomes are often only hidden in the publications." A targeted search to compare one's own data or to compare individual genes with other species is time-consuming. Although there have already been attempts to make the published genome data accessible by means of text mining, "this works so-so," says Usadel. Even the established so-called BLAST algorithm, which filters out matches in a genome for short DNA sequences, for example, only provides a limited gain in information - if you even find the comparative genome in the publications.
Jülich's individual project, funded by the Federal Ministry of Education and Research from May 2013 to December 2016 with 873,640 euros, has improved all this. "We already had a database before," says Usadel, "but its maintenance was expensive manual work." The new database also makes use of open source software. "This is cheaper and allows us to intervene directly in the code if necessary," emphasizes the bioinformatician. A linked content management system makes it easy to maintain and, above all, use the data, including graphic processing. The real highlight, however, is that a large part of the processing of new genome data can be automated.
More than 200 annotated genomes in the database
The way there was at first still laborious. The researchers had to compile the published genomes from the various publications. The information from more than 200 publications has since been entered into the database. In the beginning, these genomes had to be annotated by hand: Which genes are there, what function do they have? But that's not all: "We combine genomic and transcriptomic plant data and the associated phenotypes," explains Usadel. This is a bit like not only being able to check which sentences appear in a book, but also knowing the order of the sentences, who reads them out and what they mean.
The software provides an overview of the sequenced and published genomes.
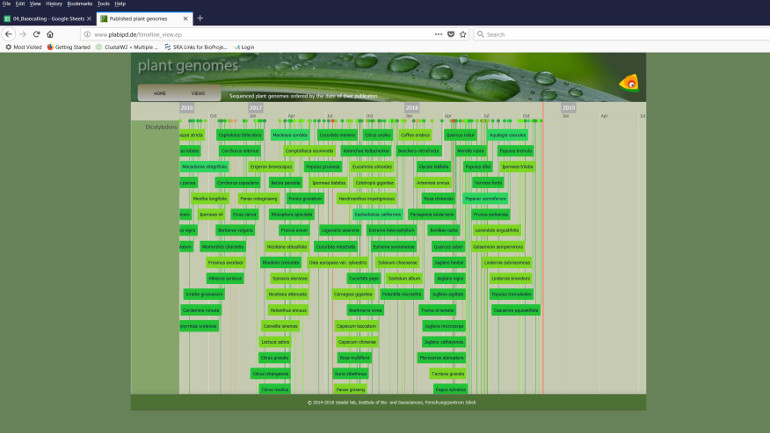
In order to enable a high degree of automation of this work, Jülich scientists are developing a standardized format for the description of phenotypes with colleagues from all over the world, driven by the phenotyping projects IPPN and EMPHASIS. Their hope is that this format will always be used in the future, as it will allow the information in the database to be automatically linked to genotypes.
New annotations automatically in minutes
A learning algorithm helps to provide new genomes with extensive annotations by comparing the new data with already annotated genomes and drawing conclusions about the significance of the new sequences. "We have already done very good annotations on new genomes," said Usadel. The sequences of the genomes in the database are already 40 to 60 percent annotated. "When we read in a new genome, we get a very clear, formal annotation with very good precision after a few minutes."
The visualization is also helpful for researchers who use the publicly accessible database. Genes are divided into groups and subgroups according to their function. Regulatory relationships between genes and their products are linked to these and to each other in a kind of schematic. "Many groups have created a transcriptome and then want to know: What does that matter at all?" Usadel explains the benefit. Where the phenotype is known, it is also linked directly - for example, whether a gene is relevant for disease resistance.
Demand greater than expected
In general, the work continues steadily after the end of project funding. "That was also a funding requirement," says Usadel. "Otherwise, the database would soon become obsolete and useless. His team is constantly reading publications so as not to miss new genomes and to improve database annotations. This work is financed by Forschungszentrum Jülich.
The effort pays off: "We have many individual queries in our database every day and have also used this for our single molecule sequencing using nanopore technology," said the project leader. Some of the associated evaluation tools have already been quoted more than 7,000 times by Google Scholar. "We knew that the need for good annotations was great. But we were surprised by the scale of the demand."
Autor: Björn Lohmann


