Visualising DNA sections in living plant cells
Plant molecular biolologists from Gatersleben and Kaiserslautern use the CRISPR-Cas system to visualise genomic sections and to decipher their functions.
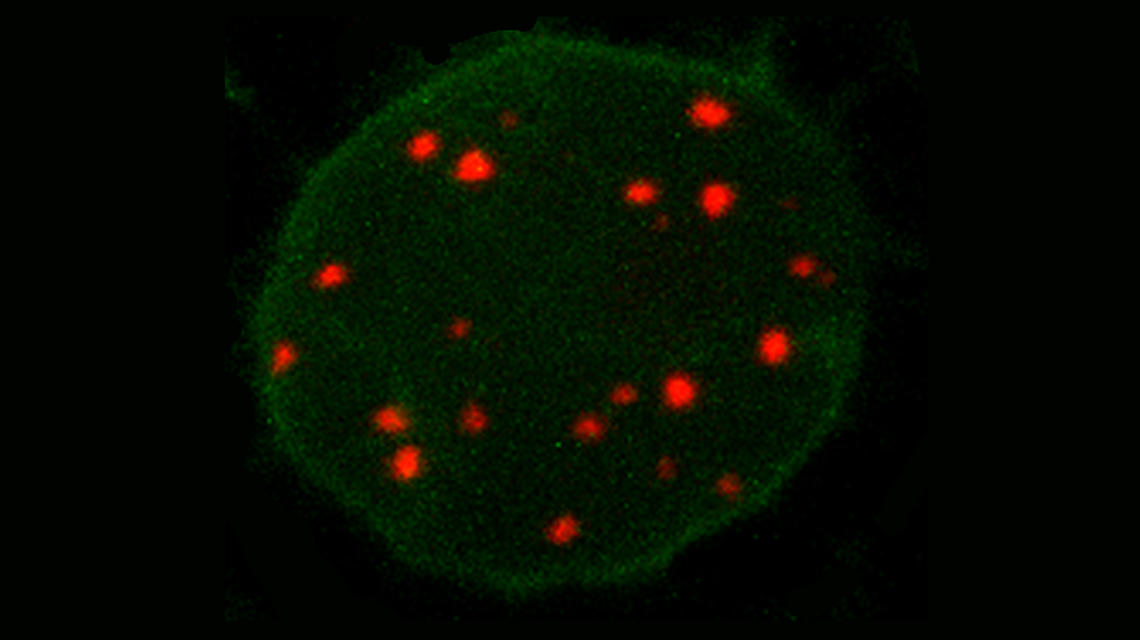
Over the last five years the new CRISPR-Cas genome editing tool has revolutionized molecular biology. The new technique allows for completely new ways of genetic engineering with relatively little effort at all. Based on this method a team of researchers headed by Andreas Houben at the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) in Gatersleben and Holger Puchta from the Botanical Institute of the Karlsruhe Institute of Technology developed a method to visualize defined genomic sequences in living plant cells. With this new technique they were able to visualise dynamic movements of chromosome ends.
Visualizing several gene loci
“By harnessing this system for live cell imaging in plants, our team shows the potential of this technology reaches far beyond the controlled induction of mutations”, explains Steven Dreissig from the IPK. “We demonstrate reliable imaging of telomere repeats located at the ends of the chromosome arms in living cells of Nicotiana benthamiana, a close Australian relative of tobacco, and pave the way for potential visualization of multiple genomic loci.” The researchers published their results in "The Plant Journal".
Organisation reflects function
This new method enables the analysis of the spatio-temporal organisation of the genome. In turn, understanding the organisation could hold the key to understanding how genome structure and function are intertwined. The spatial and temporal organization of genomes is especially important for maintaining and regulating important cell functions such as gene expression and DNA repair. Thus, elucidating the spatio-temporal organisation of the genome within the nucleus is imperative in order to understand how genes and non-coding DNA sequences are regulated during development.
Visualising DNA and proteins at once
Mapping the functional organization of the genome can be achieved by visualizing interactions between different genomic elements in living cells. “While fluorescence-tagged nuclear proteins can be readily imaged in living plant cells, in vivo visualization of defined DNA sequences turned out to be technically difficult”, explains Andreas Houben, head of the research group Chromosome Structure and Function of the IPK.” Holger Puchta, Director of the Botanical Institute in Karlsruhe adds: “Furthermore, we show that CRISPR-Cas can be combined with fluorescence-labelled proteins to investigate DNA-protein interactions in living cells.” This novel development enables scientists to visualise and thus manipulate distinct gene loci more easily in the near future.
jmr


