Better understanding of the natural function of CRISPR
Gene scissors are an important tool in genome editing, but they originated in the battle between bacteria and phages.

In 2020, the Nobel Prize in Chemistry was awarded for the discovery of the CRISPR/Cas gene scissors. The awarding of the prize took place only shortly after the actual date of discovery - a sign of high relevance. The gene scissors make it possible to edit genetic material with extreme precision. Applications range from the treatment of hereditary diseases to the rapid adaptation of important food crops to climate change. However, CRISPR in its original form is not a process devised by humans, but a natural mechanism. A team of researchers at the University of Bonn has now studied this mechanism and gained new insights.
Phage genome scissors
In nature, CRISPR is a function of the bacterial immune system, which serves to defend against phages. Phages are to bacteria what viruses are to us humans. CRISPR detects when phages attack the bacterial cell and cuts off the invader's genetic material. If the phage could integrate its genetic material intact into the bacterial genome, the bacterium would automatically use it to produce new phages and spread the infection. To be better prepared and to be able to recognize phages even faster, the bacterium stores the fragments of the phage genetic material in a kind of archive.
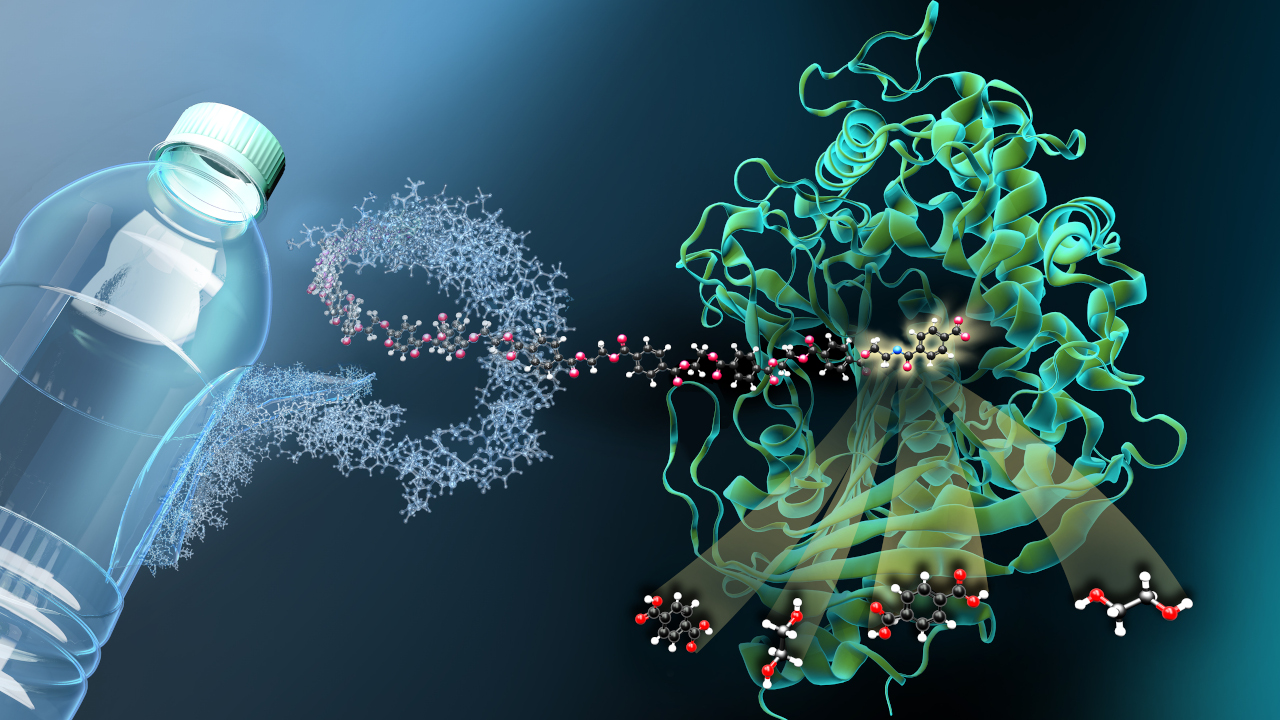
Up until this step, the natural function of CRISPR is well known. Just like the fact that the so-called type III variant of the gene scissors produces small signal molecules when it discovers the genetic material of phages. These signal molecules mobilize the bacterium's entire immune defenses to repel the attack as broadly as possible. Researchers at the University of Bonn, together with teams from the University of St. Andrews in Scotland and the European Molecular Biology Laboratory in Hamburg, have now described this signaling mechanism in more detail for the first time.
Signaling cascade for broad immune response
As the biologists report in the journal Nature, the signal molecule binds to a protein called CalpL and thus has the function of an enzyme from the group of proteases. The name comes from the fact that they can degrade certain proteins. But that's not all: "Proteases are also used in the human immune system to pass on information at a rapid pace," says Niels Schneberger of the University Hospital Bonn and one of the two first authors of the study.
Further investigations led to the target protein of the protease: CalpT. It "secures" a third protein, CalpS. If CalpT precipitates because the protease cuts it, CalpS can exert its effect. "CalpS is a well-guarded sigma factor," explains Christophe Rouillon, a visiting scientist at the university hospital and another first author of the study. "Such proteins lead to the switching on of specific genes, which sets the bacterium's metabolism to defense." Exactly which genes these are is now the subject of further research.
bl